|
Training stages abroad
I. Stage abroad participant: Dr. Ibanescu Sorin
Practical mobility QCM-D Optimization Course organized by Biolin Scientific, Gothenburg, Sweden
Period: 04.05 – 10.05. 2018
During the period 4.05.2018 - 10.05.2018, I went to Gothenburg, Sweden, to participate to a quartz crystal microbalance with dissipation (QCM-D) optimization course organized by Biolin Scientific, the world biggest QCM-D manufacturer.
 During the training I learn theory and get hands-on training in the lab - to optimize your skills on how to handle your instrument and work with Dfind to extract maximum information from your raw data. The training agenda included: QCM-D applications and introduction to additional modules; Generating quality data – Optimising experimental conditions; Hands-on lab session: Protein adsorption using Q-Sense Analyzer; Introduction to QCM-D software and data analysis; Data analysis with Dfind - Dfind demo - Dfind hands-on session During the training I learn theory and get hands-on training in the lab - to optimize your skills on how to handle your instrument and work with Dfind to extract maximum information from your raw data. The training agenda included: QCM-D applications and introduction to additional modules; Generating quality data – Optimising experimental conditions; Hands-on lab session: Protein adsorption using Q-Sense Analyzer; Introduction to QCM-D software and data analysis; Data analysis with Dfind - Dfind demo - Dfind hands-on session
There was also a Consultation Session - where I got the opportunity to discuss your personal experimental set-up and data handling in a group session with one of our QCM-D specialists. There, I got the opportunity to discuss my personal experimental set-up and data handling in a group session with Biolin QCM-D specialists. Overall, the whole experience added to my understanding of QCM-D. During that period I had the opportunity to visit the surrounding areas and meet with specialists from Biolin in a less formal setting.
II. Stage abroad participant: Dr. Andrei Neamtu
Practical mobility Participation at International Symposium & Summer School, Hochschule
Rhein Main, Russelsheim, Germany
Period: August 19 – 25th 2018
 The purpose of this stage was the participation, as an invited speaker at the International Symposium & Summer School, Mass spectrometry in Medical Technology and Biotechnology held in Russelsheim in August 2018. This summer school encompass newest technologies, centred around mass spectrometry (MS) in biotechnology and clinical applications, particularly diagnostics and new therapeutic approaches based on antibodies and antibody epitopes; biochemical applications of affinity techniques; combinations of MS and biosensor tools for affinity determination and quantification; Identification of antigen epitopes and antibody paratopes; clinical applications of affinity- proteomics; new developments in medical imaging technologies. These technologies are gaining increased importance for many application areas in pharmacology, clinical diagnostics and the development of antibodybased drugs. The Workshop and Summer School program comprised lectures, seminars & tutorials in these areas, and several practicals, performed in the Steinbeis Centre for Biopolymer Analysis. The purpose of this stage was the participation, as an invited speaker at the International Symposium & Summer School, Mass spectrometry in Medical Technology and Biotechnology held in Russelsheim in August 2018. This summer school encompass newest technologies, centred around mass spectrometry (MS) in biotechnology and clinical applications, particularly diagnostics and new therapeutic approaches based on antibodies and antibody epitopes; biochemical applications of affinity techniques; combinations of MS and biosensor tools for affinity determination and quantification; Identification of antigen epitopes and antibody paratopes; clinical applications of affinity- proteomics; new developments in medical imaging technologies. These technologies are gaining increased importance for many application areas in pharmacology, clinical diagnostics and the development of antibodybased drugs. The Workshop and Summer School program comprised lectures, seminars & tutorials in these areas, and several practicals, performed in the Steinbeis Centre for Biopolymer Analysis.
My lecture had the title “In silico Epitope Mapping Using Replica Exchange Molecular Dynamics (REMD) Simulations”. I also participated in the summer school teaching activities at Hochschule Rhein Main where the main topics included antibody characterization, peptide and protein epitope identification, new developments in protein therapy, clinical diagnostics, biomarker discovery, protein aptamers and biosensors and affinity-mass spectrometry. I also participated in the summer school teaching activities at Hochschule Rhein Main where the main topics included antibody characterization, peptide and protein epitope identification, new developments in protein therapy, clinical diagnostics, biomarker discovery, protein aptamers and biosensors and affinity-mass spectrometry.
III. Stage abroad participant: Dr. Irina Rosca
Practical mobility Institute of Biotechnology INBIOTEC Leon, Spania
Period: 20.09 – 10.10.2018
 The mobility was from September 20th to October 10th at the Institute of Biotechnology INBIOTEC, Leon, Spain and aimed to generate economically feasible technologies and environmentally friendly, based on biology synthetic, to increase the life of the wood used in interior or exterior fittings against degradation. The wood samples tested at INBIOTEC come from the Romanian and Spanish forests, and at PPIMC, Iași where was donte the synthesis and development of biocoatings based on functionalised vegetable oils and the evaluation of parameters such as: brightness, color , cracks, for wood protected with biocoatings. These wood samples were tested at INBIOTEC, Leon, where the following types of activities were carried out: Covering the pine wood samples (with Romanian and Spanish origin) with 5 types of functionalized vegetable; covering pine wood samples with 5 types of functionalized vegetable oils improved with 3 types of spores isolated from microorganisms (Bacillus sp., Streptomyces sp., Trichoderma sp.) in a concentration of 1 x 106 in order to amplify the activity protection provided by functionalized vegetable oils; parallel testing of all types of coatings together with untreated controls: testing the resistance to the activity of fungal strains (A. brasiliensis, C. puteana, T. diversicolor) by incubation in PDA (potato dextrose agar) medium at 25ºC for 8 weeks. isolation of bacterial and fungal DNA, as well as of spores produced by various micro-organisms (Bacillus sp, Trichoderma sp, Streptomyces sp) in order to identify microorganisms that attack the wood and to use their spores in the fight against wood degradation; The mobility was from September 20th to October 10th at the Institute of Biotechnology INBIOTEC, Leon, Spain and aimed to generate economically feasible technologies and environmentally friendly, based on biology synthetic, to increase the life of the wood used in interior or exterior fittings against degradation. The wood samples tested at INBIOTEC come from the Romanian and Spanish forests, and at PPIMC, Iași where was donte the synthesis and development of biocoatings based on functionalised vegetable oils and the evaluation of parameters such as: brightness, color , cracks, for wood protected with biocoatings. These wood samples were tested at INBIOTEC, Leon, where the following types of activities were carried out: Covering the pine wood samples (with Romanian and Spanish origin) with 5 types of functionalized vegetable; covering pine wood samples with 5 types of functionalized vegetable oils improved with 3 types of spores isolated from microorganisms (Bacillus sp., Streptomyces sp., Trichoderma sp.) in a concentration of 1 x 106 in order to amplify the activity protection provided by functionalized vegetable oils; parallel testing of all types of coatings together with untreated controls: testing the resistance to the activity of fungal strains (A. brasiliensis, C. puteana, T. diversicolor) by incubation in PDA (potato dextrose agar) medium at 25ºC for 8 weeks. isolation of bacterial and fungal DNA, as well as of spores produced by various micro-organisms (Bacillus sp, Trichoderma sp, Streptomyces sp) in order to identify microorganisms that attack the wood and to use their spores in the fight against wood degradation;
IV. Stage abroad participant: Olaru Craciun Anda Mihaela
Practical mobility Florence University; Neurofarba Department, Section of Pharmaceutical
and Nutriceutical Sciences, Florence, Italy
Period: July 8th – September 9th 2019
 Our target was to synthesis a series of phenothiazine based compounds with a proper design for different isoform of carbonic anhydrase inhibition. These compound are based on imine linkage and, respectively, a reduced imine linkage (corresponding compounds with amine group). All the obtained compounds were fully characterized from structural point of view. In this months, together with Luminita Marin, the synthesis protocol was developed and in this way were synthetized 4 compounds. This study will be continued in order to obtain all compounds (7) which will be tested on carbon anhydrase. After fully characterization and interpretation data, from structural, biological and structure-activity relationship point of view, the obtained results will be published as scientific article. In this period, our work in the laboratory was very pleasant due to very nice people, extremely dedicated to the research activity and their help to have an enjoyable period in order to finish the research activity, but also to visit the beautiful places in Italy. Our target was to synthesis a series of phenothiazine based compounds with a proper design for different isoform of carbonic anhydrase inhibition. These compound are based on imine linkage and, respectively, a reduced imine linkage (corresponding compounds with amine group). All the obtained compounds were fully characterized from structural point of view. In this months, together with Luminita Marin, the synthesis protocol was developed and in this way were synthetized 4 compounds. This study will be continued in order to obtain all compounds (7) which will be tested on carbon anhydrase. After fully characterization and interpretation data, from structural, biological and structure-activity relationship point of view, the obtained results will be published as scientific article. In this period, our work in the laboratory was very pleasant due to very nice people, extremely dedicated to the research activity and their help to have an enjoyable period in order to finish the research activity, but also to visit the beautiful places in Italy.
After two months, dr. Fabrizio Carta proposed us to come back in Florence as soon as possible in order to continue our research and also to start another field on their research area, on carbon anhydrase inhibition.
V. Stage abroad participant: Craciun Bogdan Florin
Practical mobility at Florence University; Neurofarba Department, Section of
Pharmaceutical and Nutriceutical Sciences, Florence, Italy
Period: July 8th – September 9th 2019
 Our research plan for this collaborative mobility was to obtain some squalene based derivatives with coumarines and sulfonamides and test their inhibition activity on different isoforms of carbon anhydrase (CA). Based on this plan, together with Lilia Clima, we’ve managed to obtain 5 squalene derivatives (2 coumarines and 3 sulfonamides) which were fully characterized from structural point of view and tested on CA I, II, IX and XII isoforms. The data obtained from CA testings showed that one of the compounds with sulfonamide presented very good results on CA II isoform and these results will be published very soon after we have all the required data. The stay in Florence was very pleasant and beneficial for scientific purpose. The people in the laboratory were very nice, extremely noble-minded and dedicated to their research activities. During our stay in Florence we were able to visit very beautiful places and to eat very delicious Italian food. Regarding professor Claudiu Supuran which is the head of the laboratory at the Florence University, we meet him several times because he is very busy and travel a lot. He is an incredible person and helped us a lot with everything during this period and I have noticed that his group have a lot of respect for him and they are working very hard to make him proud. At the end of the practical mobility, dr. Fabrizio Carta, who is professor Supuran most trusted person, confessed his desire to come back in Florence as soon as possible to continue our research. In conclusion, these 2 months period was very productive both scientifically and collaboratively in the ongoing and future projects. The people in the laboratory were very kind and they helped us with everything. The laboratory is well organized and equipped having all the materials, solvents and reagents needed for all types of synthesis. Our research plan for this collaborative mobility was to obtain some squalene based derivatives with coumarines and sulfonamides and test their inhibition activity on different isoforms of carbon anhydrase (CA). Based on this plan, together with Lilia Clima, we’ve managed to obtain 5 squalene derivatives (2 coumarines and 3 sulfonamides) which were fully characterized from structural point of view and tested on CA I, II, IX and XII isoforms. The data obtained from CA testings showed that one of the compounds with sulfonamide presented very good results on CA II isoform and these results will be published very soon after we have all the required data. The stay in Florence was very pleasant and beneficial for scientific purpose. The people in the laboratory were very nice, extremely noble-minded and dedicated to their research activities. During our stay in Florence we were able to visit very beautiful places and to eat very delicious Italian food. Regarding professor Claudiu Supuran which is the head of the laboratory at the Florence University, we meet him several times because he is very busy and travel a lot. He is an incredible person and helped us a lot with everything during this period and I have noticed that his group have a lot of respect for him and they are working very hard to make him proud. At the end of the practical mobility, dr. Fabrizio Carta, who is professor Supuran most trusted person, confessed his desire to come back in Florence as soon as possible to continue our research. In conclusion, these 2 months period was very productive both scientifically and collaboratively in the ongoing and future projects. The people in the laboratory were very kind and they helped us with everything. The laboratory is well organized and equipped having all the materials, solvents and reagents needed for all types of synthesis.
VI. Stage abroad participant: Isac Dragos Lucian
Practical mobility at Scientific Summer School "Molecular Modeling: Real Applications and
New Approaches - Pula, Sardinia – Italia
Period: 29 July - 2 August 2019
 The School was targeted at students, graduate students, early-stage researchers, researchers, technologists and professionals in sectors as biomedicine, nutraceuticals, functional foods, physics, biology, chemistry, biochemistry, materials science, nanosciences, energy and environmental sciences. The courses focused on how to apply molecular modeling in different fields such as chemistry, physics, biology, and materials. As follows different levels of theory (based on quantum mechanics, quantum mechanics/molecular mechanics, molecular mechanics, molecular dynamics) were presented at these courses. The courses have been included the lectures from the electronic structure up to life systems. The courses were followed by tutorials in order to understand how to apply the methods. The School was targeted at students, graduate students, early-stage researchers, researchers, technologists and professionals in sectors as biomedicine, nutraceuticals, functional foods, physics, biology, chemistry, biochemistry, materials science, nanosciences, energy and environmental sciences. The courses focused on how to apply molecular modeling in different fields such as chemistry, physics, biology, and materials. As follows different levels of theory (based on quantum mechanics, quantum mechanics/molecular mechanics, molecular mechanics, molecular dynamics) were presented at these courses. The courses have been included the lectures from the electronic structure up to life systems. The courses were followed by tutorials in order to understand how to apply the methods.
Therefore, many students have been applied for this school. The lectures were presented by different professors with background in molecular modeling (Dario Estrin, Aatto Laaksonen, Amit Kumar, Santiago Di Lella, Zhongyuan Lu, Francesco Sciortino, Francesca Mocci, Giancarlo Cappellini, M. Natalia Dias Soares Cordeiro, Claudio Melis, Roberto Cardia, Maria Valentini, Vincenzo Martorana, Enrico Pieroni.
https://www.biolinscientific.com/
VII. Stage abroad participant: Vasiliu Tudor
Practical mobility at Scientific Summer School "Molecular Modeling: Real Applications and
New Approaches - Pula, Sardinia – Italia
and Cagliari University, Italy
Period: 28 June - 2 August 2019
 The objective of the mobility to Cagliari University was the development of a Course Grain model that would allow the simulation of a huge non-viral vector. During my stay there I worked in the group of Dr. Francesca Mocci where I had the opportunity to learn from researchers experienced in Molecular Modeling. Using the Magic Software and the Inverse-Monte Carlo method we obtained a preliminary model for the vector. Due to the complexity and size of the vector further optimization of the model needs to be done in order to mimic the real life interactions of the vector with the DNA. Also during my stay in Italy I attended the Scientific Summer School "Molecular Modeling: Real Applications and New Approaches" 29th July- 2nd August 2019 - Pula, Sardinia – Italy. During the summer school I had the opportunity to interact with some of the lead scientists in Molecular Modeling (like professor Aatto Larksonen or professor Zhongyuan Lu) and I was able to learn new techniques and methods used in Molecular Modeling Simulations. The school was also a good opportunity to get a sense of the current trends in molecular modeling research and learn how to disseminate in an efficient way my research. The objective of the mobility to Cagliari University was the development of a Course Grain model that would allow the simulation of a huge non-viral vector. During my stay there I worked in the group of Dr. Francesca Mocci where I had the opportunity to learn from researchers experienced in Molecular Modeling. Using the Magic Software and the Inverse-Monte Carlo method we obtained a preliminary model for the vector. Due to the complexity and size of the vector further optimization of the model needs to be done in order to mimic the real life interactions of the vector with the DNA. Also during my stay in Italy I attended the Scientific Summer School "Molecular Modeling: Real Applications and New Approaches" 29th July- 2nd August 2019 - Pula, Sardinia – Italy. During the summer school I had the opportunity to interact with some of the lead scientists in Molecular Modeling (like professor Aatto Larksonen or professor Zhongyuan Lu) and I was able to learn new techniques and methods used in Molecular Modeling Simulations. The school was also a good opportunity to get a sense of the current trends in molecular modeling research and learn how to disseminate in an efficient way my research.
VIII. Stage abroad participant: Dr. Lilia Clima
Practical mobility at Florence University; Neurofarba Department, Section of
Pharmaceutical and Nutriceutical Sciences, Florence, Italy
Period: 17 June – 10 July 2019
The main objective of my travel to University of Florence to Claudiu Supuran’s group, was to design synthesize ant test a series of compounds as inhibitors for carbonic anhydrase. The syntheses focused on the functionalization of the squalene with sulphonamide or coumarin groups as presented below:
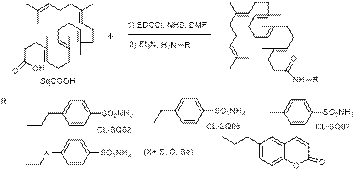
 SqCOOH was obtained in 4 step synthesis as described by us in the literature (B. Craciun, T. Vasiliu, N. Marangoci, M. Pinteala, L. Clima, Rev. Roum. Chim. 2018, 63, 621-628), activated as N-hydroxysuccinimide ester with EDCCl in DMF and coupled with amines (H2N-R) in second step to obtain functionalized amides.From proposed reaction two compounds, CL-SQ02 and CL-SQ06 were obtained and characterized. The other reactions did not lead to the desire products or the products couldn’t be purified. Compound Cl-SQ02 showe a significant inhibition ofcarbonoc anhydrase hCAII species. SqCOOH was obtained in 4 step synthesis as described by us in the literature (B. Craciun, T. Vasiliu, N. Marangoci, M. Pinteala, L. Clima, Rev. Roum. Chim. 2018, 63, 621-628), activated as N-hydroxysuccinimide ester with EDCCl in DMF and coupled with amines (H2N-R) in second step to obtain functionalized amides.From proposed reaction two compounds, CL-SQ02 and CL-SQ06 were obtained and characterized. The other reactions did not lead to the desire products or the products couldn’t be purified. Compound Cl-SQ02 showe a significant inhibition ofcarbonoc anhydrase hCAII species.
During the research stay, I tightly collaborated with Claudiu Supuran and Fabrizio Carta, discussing the design strategies for the preparation of compounds. In the lab, Andrea Angeli and Murat Bozdag supported me. From my opinion, a tight collaboration has started originating connected research fields.
IX. Stage abroad participant: Dr. Luminita Marin
Practical mobility at Florence University; Neurofarba Department, Section of
Pharmaceutical and Nutriceutical Sciences, Florence, Italy
Period: 17 June – 10 July 2019
 The stage has the aim to establish together with Professor Claudiu Supuran, a new research direction consisting in the synthesis and characterization of new compounds for carbonic anhydrase inhibition, targeting anticancer properties. Carbonic anhydrases (CA, EC 4.2.1.1) catalyze the interconversion bewteen carbon dioxide and bicarbonate with generation of protons. The carbonic anhydrase isozyme IX (CA IX) is highly overexpresed in hypoxic tumors and shows very restricted expression in normal tissues. CA IX is a dimeric protein possessing very high catalytic activity for the hydration of carbon dioxide to protons and bicarbonate. Its quaternary structure is unique among members of this family of enzymes, allowing for structure-based drug design campaigns of selective inhibitors. In this line of thought we designed a series of phenothiazine-based sulphonamides, and during my stage I started their synthesis and purification, my work being continued by my co-worker Dr. Anda-Mihaela Craciun. Besides the synthesis of new anticancer compounds, I performed a large documentation activity on this new research direction, I learnt a new technique consisting in measurements of carbonic anhydrase inhibition and I established good collaboration relationship with the team members of the Professor Supuran. Overall, the stage constituted a good opportunity to enrich the activity of my team with a new research direction and a new scientific colaboration, which can be a starting point for new research projects. The stage has the aim to establish together with Professor Claudiu Supuran, a new research direction consisting in the synthesis and characterization of new compounds for carbonic anhydrase inhibition, targeting anticancer properties. Carbonic anhydrases (CA, EC 4.2.1.1) catalyze the interconversion bewteen carbon dioxide and bicarbonate with generation of protons. The carbonic anhydrase isozyme IX (CA IX) is highly overexpresed in hypoxic tumors and shows very restricted expression in normal tissues. CA IX is a dimeric protein possessing very high catalytic activity for the hydration of carbon dioxide to protons and bicarbonate. Its quaternary structure is unique among members of this family of enzymes, allowing for structure-based drug design campaigns of selective inhibitors. In this line of thought we designed a series of phenothiazine-based sulphonamides, and during my stage I started their synthesis and purification, my work being continued by my co-worker Dr. Anda-Mihaela Craciun. Besides the synthesis of new anticancer compounds, I performed a large documentation activity on this new research direction, I learnt a new technique consisting in measurements of carbonic anhydrase inhibition and I established good collaboration relationship with the team members of the Professor Supuran. Overall, the stage constituted a good opportunity to enrich the activity of my team with a new research direction and a new scientific colaboration, which can be a starting point for new research projects.
X. Stage abroad participant: Dr. Dragos Pieptanariu
Practical mobility at Florence University; Neurofarba Department, Section of
Pharmaceutical and Nutriceutical Sciences, Florence, Italy
Period: August 26 and September 9, 2019
 Our medical doctor Dragos Peptanariu, research scientist in our group, went for a two-week research internship in the group of Prof. Claudiu T. Supuran, at the University of Florence, Italy between August 26 and September 9, 2019. Through this internship Mr. Peptanariu met collaborators of Prof. Claudiu Supuran, among whom the research group of Dr. Lorenzo Di Cesare Mannelli from the Department of Neuroscience, Psychology, Drug Research and Child Health - NEUROFARBA from Florence. His stage was focus on the inhibition of CA IX with sulfonamide and/or coumarin inhibitors that was recently shown to lead to a potent retardation for the growth of both primary tumors and metastases. Some fluorescent sulfonamides were shown to accumulate only in hypoxic tumor cells overexpressing CA IX, and might be used as diagnostic tools for imaging of hypoxic cancers. During this visit they exchanged knowledge in the field of in vitro biological tests and Mr. Peptanariu had the opportunity to observe certain techniques such as separation of mononuclear cells from the blood, or preparation of rat spinal cord sections for microscopy and he also met Doctor Jose' Manuel Pioner from Dipartimento di Medicina Sperimentale e Clinica, University of Florence. Mr. Peptanariu and Mr. Pioner talked about induced pluripotent stem cells and their differentiation into cardiomyocytes. Our medical doctor Dragos Peptanariu, research scientist in our group, went for a two-week research internship in the group of Prof. Claudiu T. Supuran, at the University of Florence, Italy between August 26 and September 9, 2019. Through this internship Mr. Peptanariu met collaborators of Prof. Claudiu Supuran, among whom the research group of Dr. Lorenzo Di Cesare Mannelli from the Department of Neuroscience, Psychology, Drug Research and Child Health - NEUROFARBA from Florence. His stage was focus on the inhibition of CA IX with sulfonamide and/or coumarin inhibitors that was recently shown to lead to a potent retardation for the growth of both primary tumors and metastases. Some fluorescent sulfonamides were shown to accumulate only in hypoxic tumor cells overexpressing CA IX, and might be used as diagnostic tools for imaging of hypoxic cancers. During this visit they exchanged knowledge in the field of in vitro biological tests and Mr. Peptanariu had the opportunity to observe certain techniques such as separation of mononuclear cells from the blood, or preparation of rat spinal cord sections for microscopy and he also met Doctor Jose' Manuel Pioner from Dipartimento di Medicina Sperimentale e Clinica, University of Florence. Mr. Peptanariu and Mr. Pioner talked about induced pluripotent stem cells and their differentiation into cardiomyocytes.
XI. Stage abroad participant: Dr. Irina Rosca
Practical mobility Institute of BiotechnologyINBIOTECH, Leon, Spania
Period: 18.11 – 08.12. 2019
The main objective of the mobility project held from November 18th to December 8th at the Institute of Biotechnology INBIOTEC, Leon, Spain, under the coordination of the director Dr. Carlos Barreiro was to test of a economically feasible and environmentally friendly product, synthesized at the Petru Poni Institute of Macromolecular Chemistry from Iași (PPIMC) based on synthetic biology, aiming at increasing the wood lifetime by combating the fungal degradation. In this context, a range of coatings based on functionalized vegetable oils were synthesized within the group of the IntelCentru department of PPIMC, of which, based on preliminary selection the most promising one in terms of composition and deposition mode on wood surface was chosen. The wood samples tested at INBIOTEC come from the Romanian forests, and at PPIMC, Iași, was done the synthesis and development of coatings based on vegetable oils, their functionalization and the evaluation of wood parameters such as: brightness, color and cracks. These wood samples were tested at INBIOTEC, Leon, where the following types of activities were performed: coating the wood samples with an extract of eucalypt wood with antimicrobial and antioxidant activity used as a primer for the biocoating; coating the pine wood samples with the new synthetized compound produced in PPIMC; coating the wood samples with the biocoating improved with bacterial spores isolated in INBIOTEC in order to improve the antifungal activity of the functionalized oils; parallel testing of the antifungal activity of the coatings together with untreated wood samples against fungal strains isolated and characterized in INBIOTEC (Aspergillus brasiliensis, Coniophora puteana, Trametes verisicolor) by incubating in potato dextrose agar medium (PDA) at 25±1°C for 10 weeks. The resulted samples will be send to PPIMC for physical and chemical characterization. Other activities carried out during the research internship included: isolation of bacterial and fungal DNA, bacterial proteins, as well as of spores produced by various microorganisms in order to identify those that attack the fungal strains that produce wood damage. |



